|
|
Discovery There is astonishing chemical diversity among RNA modifications. However, identifying the complement of modifications in specific types or pools of RNA, particularly less abundant RNAs such as mRNAs, has been challenging. We use mass spectrometry (LC-MS/MS) to discover modifications in RNAs of interest, and follow up with mechanistic studies (below). LC-MS/MS is also a powerful tool to quantify modifications under varying conditions. |
|
|
Characterization To begin to uncover the cellular functions of modifications on RNA, we map their locations using high throughput sequencing and identify their regulatory proteins. When necessary (and it often is), we develop new methods to achieve this. Once we understand where these modifications occur and which proteins install, remove, and bind them, we are well-equipped to identify what their cellular functions may be. |
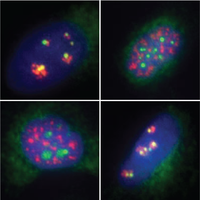 |
Mechanism We use a variety of methods understand how RNA modifications are involved in cellular processes, including biochemistry, microscopy, reporter assays, proteomics, high throughput sequencing and more. Our goal is two-fold: to understand the mechanisms by which RNA modifications impact cell signaling, and to identify the processes in which these mechanisms are the most relevant. The biology always guides our choice of technique and model system. |
 |
Application As a result of their core functions in gene expression regulation, modifications on many types of RNA have been connected to numerous human diseases. We always seek to connect our work to human development and disease, and try to identify how our findings may open novel therapeutic avenues. |